More Information
Submitted: February 20, 2021 | Approved: February 24, 2021 | Published: February 25, 2021
How to cite this article: Wang SY, Lin LT, Lin BZ, Chang CH, Chang CY, et al. A comparative study of single or dual treatment of theranostic 188Re-Liposome on microRNA expressive profiles of orthotopic human head and neck tumor model. Heighpubs Otolaryngol Rhinol. 2021; 5: 001-012.
DOI: 10.29328/journal.hor.1001024
Copyright License: © 2021 Wang SY, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: 188Re-liposome; HNSCC; Cerenkov luminescent imaging; microRNA expressive profile; prognostic factors
A comparative study of single or dual treatment of theranostic 188Re-Liposome on microRNA expressive profiles of orthotopic human head and neck tumor model
Shan-Ying Wang1,2, Liang-Ting Lin3, Bing-Ze Lin4, Chih-Hsien Chang1,4, Chun-Yuan Chang1, Min-Ying Lin1 and Yi-Jang Lee1,5*
1Department of Biomedical Imaging and Radiological Sciences, National Yang Ming Chiao Tung University, School of Biomedical Engineering, 112 Taipei, Taiwan
2Department of Nuclear Medicine, Far Eastern Memorial Hospital, 220, New Taipei City, Taiwan
3Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Kowloon, Hong Kong, SAR, P.R. China
4Isotope Application Division, Institute of Nuclear Energy Research, Taoyuan, Taiwan
5Cancer Progression Research Center, National Yang-Ming University, 112, Taipei, Taiwan
*Address for Correspondence: Yi-Jang Lee, Ph.D., Department of Biomedical Imaging and Radiological Sciences, National Yang-Ming University, No. 155, Sec. 2, Linong St. Beitou District, 112, Taipei, Taiwan, Tel: +886-2-28267189; Email: [email protected]
Background: 188Re-liposome has been used for evaluating the theranostic efficacy on human head and neck squamous cell carcinoma (HNSCC) at preclinical stages. Here we furthercompared the microRNA expressive profile in orthtopic HNSCC tumor model exposed to 188Re-liposome.
Methods: A single dose or dual doses of 188Re-liposome was intravenously injected into tumor-bearing mice followed by the Cerenkov luminescent imaging (CLI) for monitoring the accumulation of 188Re-liposome in tumors. The microRNA expressive profile was generated using the Taqman® OpenArray® Human MicroRNA Panel followed by the DIANA mirPath analysis, KEGG signaling pathways prediction, and Kaplan-Meier survival analysis for predicting the prognostic role of 188Re-liposome affected microRNAs.
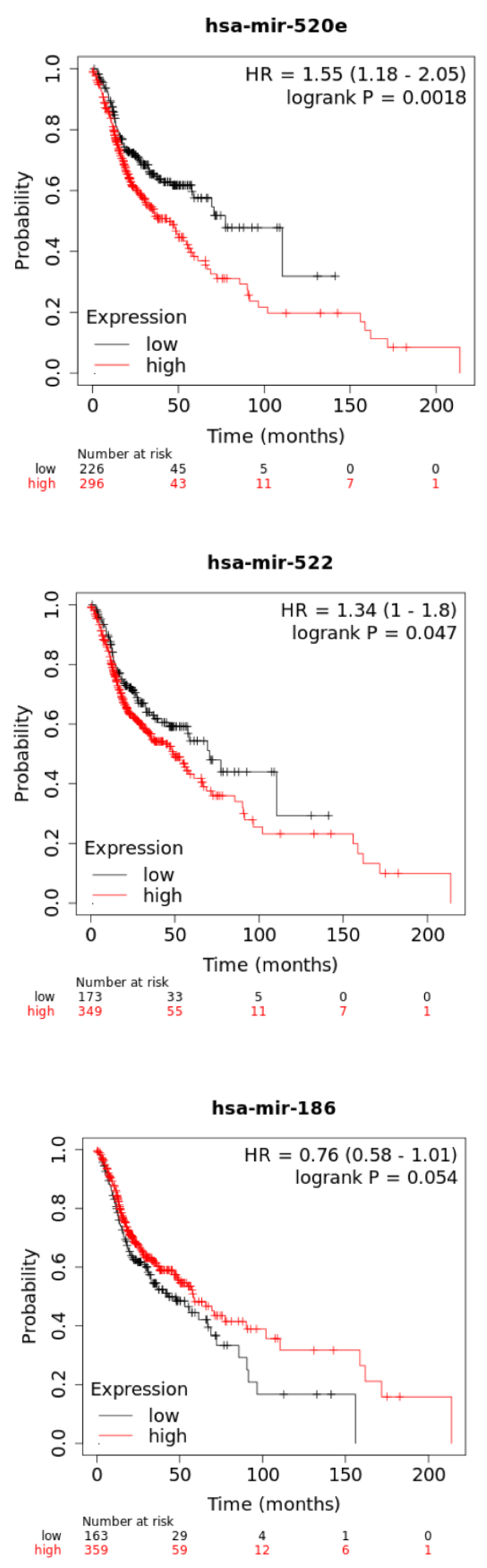
Results: Dual doses of 188Re-liposome exhibited a better tumor suppression than a single dose of 188Re-liposome, including reduced tumor size, Ki-67 proliferative marker, and epithelial-mesenchymal transition (EMT) related factors. The microRNA expressive profiles showed that 22 microRNAs and 19 microRNAs were up-regulated and down-regulated by dual doses of 188Re-liposome, respectively. Concomitantly, these two groups of microRNAs were inversely regulated by a single dose of 188Re-liposome accordingly. These microRNAs influenced most downstream genes involved in cancer related signaling pathways. Further, miR-520e and miR-522-3p were down-regulated whereas miR-186-5p and miR-543 were up-regulated by dual doses of 188Re-liposome, and they separately affected most of genes involved in their corresponding pathways with high significance. Additionally, high expressions of miR-520e and miR-522-3p were associated with lower survival rate of HNSCC patients.
Conclusion: MicroRNA expression could be used to evaluate the therapeutic efficacy and regarded prognostic factors using different doses of 188Re-liposome.
Radiogenomics is a rising field of modern applications for radiomics linking to the genetic profile of tumor progression to better predict the therapeutic responses from diagnosis to prognosis. A recent review has summarized the applications of radiogenomics in a variety of human cancers [1]. It also directly influences the gene expressive profile of target tissues because of the radiotherapy induced cytotoxicity [2]. Radiopharmaceutical is an ideal candidate for monitoring the efficacy of drug targeting and therapy using preclinical or clinical imaging modalities. However, radiopharmaceutical is little applied in radiogenomics.
Theranostics is defined as specific targeted diagnosis and therapy using a material with both effects on tumor treatment. Nuclear medicine plays an important role in theranostics as several types of radiopharmaceuticals emit both γ-rays and high energy β particles for diagnosis and therapy, respectively [3]. For instance, rhenium-188 (188Re) emits 85% of 2.12MeV β particles and 15% of 155keV γ-rays during decay, so it belongs to a theranostic radionuclide as well [4]. It is also an attractive and affordable radiopharmaceutical because of its short half-life and on-demand availability using a tungsten-188/rhenium-188 generator [4]. Moreover, the atomic radius of 188Re is similar to technetium that has been widely used in clinics [5,6]. The tissue penetration of emitted β particles is about 10 mm, suggesting that it is suitable for the treatment of large-sized or mid-late stage tumors [7]. Accumulated literature have demonstrated that liposome embedded 188Re (188Re-liposome) is able to target human colorectal cancer, glioblastomas, esophageal cancer, head and neck cancer and lung cancer using the xenograft tumor model [8-12]. Tumor accumulation of 188Re-liposome is passive on the behalf of the enhanced permeability and retention (EPR) effect, which is dependent on the mal-formation of blood vessels surrounding tumors [13,14]. Previous studies have demonstrated that 188Re-liposome could influence the gene expression in human head and neck squamous cell carcinoma (HNSCC), and the let-7 family of microRNA mediated gene expressive profile was significantly involved in this treatment [12]. Therefore, this finding intrigues us to investigate whether 188Re-liposome would also influence the microRNA expressive profile.
Although 188Re-liposome exhibited tumor accumulative property in HNSCC, the therapeutic efficacy was moderate. Specifically, regrowth of xenograft tumors were detected when they were treated by a single dose of 188Re-liposome but not by repeated doses [15]. As chemoresistance is known to be associated with a series of molecular regulation [16], we are interested in investigating the gene expressive profiles of HNSCC tumors treated with 188Re-liposome. In this study, we exploited the open arrays of microRNA to analyze over 700 microRNA in orthotopic HNSCC tumor treated with a single dose or repeated doses of 188Re-liposome. The interested microRNAs were also subjected to survival analysis. The results of 188Re-liposome modulated expression of microRNA were discussed.
Cell line
Human FaDu head and neck squamous cell carcinoma cells (American Type Culture Collection, Manassas, VA, USA) were maintained in RPMI-1640 (Life Technologies Inc., Carlsbad, CA, USA) medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamate, 50 unit/ml penicillin and 50 µg/ml streptomycin (Invitrogen, Carlsbad, CA). The pH of medium was adjusted by sodium bicarbonate. Cells were incubated at 37 oC in a humidified incubator with 5% CO2, and passaged every two days.
Preparation of Rhenium-188liposomal drug
The procedures of 188Re-liposome preparation of validation were followed as described previously [17]. The dose of each injection is 640 µCi corresponding to 80% maximum tolerated dose (MTD) [9].
HNSCC orthotopic tumor model
Four-week old male BALB/c nude mice were used to establish the orthotopic tumor model (National Laboratory Animal Center of Taiwan, Tainan, Taiwan). FaDu cells (1 x 106) were resuspended in 100 µl of OPTI-MEM and injected into the buccal positions of mice (N = 5) using a 27G insulin needle. Tumors could form about 3 weeks after tumor implantation. The study has been approved by the Institutional Animal Care and Utilization Committee (IACUC) of National Yang-Ming University (case no. 1061010).
Cerenkov luminescent imaging (CLI) and tumor resection
CLI was performed at 24 hours after the administration of 188Re-liposome. The signals were acquired by the in vivo Imaging System (IVIS 50, Perkin Elmer Inc., Waltham, MA, USA). Regions of interest (ROIs) were delineated on the tumor around the mouth. For evaluation of tumor response to 188Re-liposome, resection of tumors with various treatment were performed and compared by size.
Immunohistochemical (IHC) staining
The procedures of IHC was followed by our previous report [15]. Fixed tissue sections were incubated with anti-Ki-67 antibody (MAB4190, EMD Millipore, Billerica, MA, USA) at 4 °C overnight. The slides were rinsed and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (EnVisionTM+Dual Link System-HRP(DAB+), K4065, Dako), and developed in 3’,3’-diaminobenzidine (DAB+) substrate chromogen and counterstained with Mayer’s hematoxylin (ScyTek Laboratories, Utah, USA). The Ki-67 positivity index was quantified after the digitalization of the slides and calculated by the online ImmunoRatio automated counting program (http://153.1.200.58:8080/immunoratio/) [18].
Western blot analysis
Tumors were resected from the tumor-bearing mice after 4 weeks of 188Re-liposome treatment, and lysed using the T-PER™ Tissue Protein Extraction Reagent (Thermo Fisher Scientific, Waltham, MA) containing 1% of protease inhibitor cocktail (Sigma-Aldrich). Protein lysates were run on 8% - 12% sodium dodecyl sulfate –polyacrylamide gel electrophoresis (SDS-PAGE). The fractionated proteins were transferred based on previously [19]. The antibodies included E-cadherin (GTX100443), Slug (GTX128796), ZEB-1(GTX105278), vimentin (GTX100619), Snail (GTX100754) were from GeneTex (Genetex Inc. Irvine, CA, USA). Antibody against GAPDH (MA5-15738) was from Sigma (Sigma-Aldrich Co, St. Louis, MO, USA).
MicroRNA expressive profiling analysis
Taqman® OpenArray® Human MicroRNA Panel (Thermo Fisher Scientific Inc., Waltham, MA, USA) was used to detect the microRNA expression in orthotopic HNSCC tumors treated with different regimens of 188Re-liposome and compared to untreated control. The operation was performed in the Sequencing Core Facility of National Yang Ming University Genome Center (YMGC). For analysis, the expression levels of the two experimental groups (dual doses and single dose of 188Re-liposome) were defined by the Mean Relative Quantification (RQ) normalized to the control group. The expression level changes between the groups were demonstrated by log2 ratio. All the identified assay id of each miRNA probes was converted to miRBase ID (v22) referring to the manufacturer’s handbook. The lists were input to DIANA mirPath v.3 (http://snf-515788.vm.okeanos.grnet.gr/) for Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, referencing the DIANA-MicroT prediction [20]. The overall involvement and the co-regulated gene prediction were concluded with p - value threshold of 0.05 and MicroT threshold of 0.8 by genes union and genes intersection function, respectively.
Survival analysis for microRNA
The association of microRNA of interest with public human HNSCC microRNA database was determined by an on-line Kaplan-Meier plotter [21]. In the database, the miRNA subsystems include 11k samples from 20 different cancer types, which include 523 human HNSCC cases. The significance of microRNA associated survival rate was determined by a log-rank test.
Statistics
Data were represented as the mean ± S.D. from triplicate independent results, and the statistical analysis was performed using t-test. The statistical significance was set at p < 0.05.
Effects of single dose and dual doses of 188Re-liposome on the growth of orthotopic HNSCC tumor model
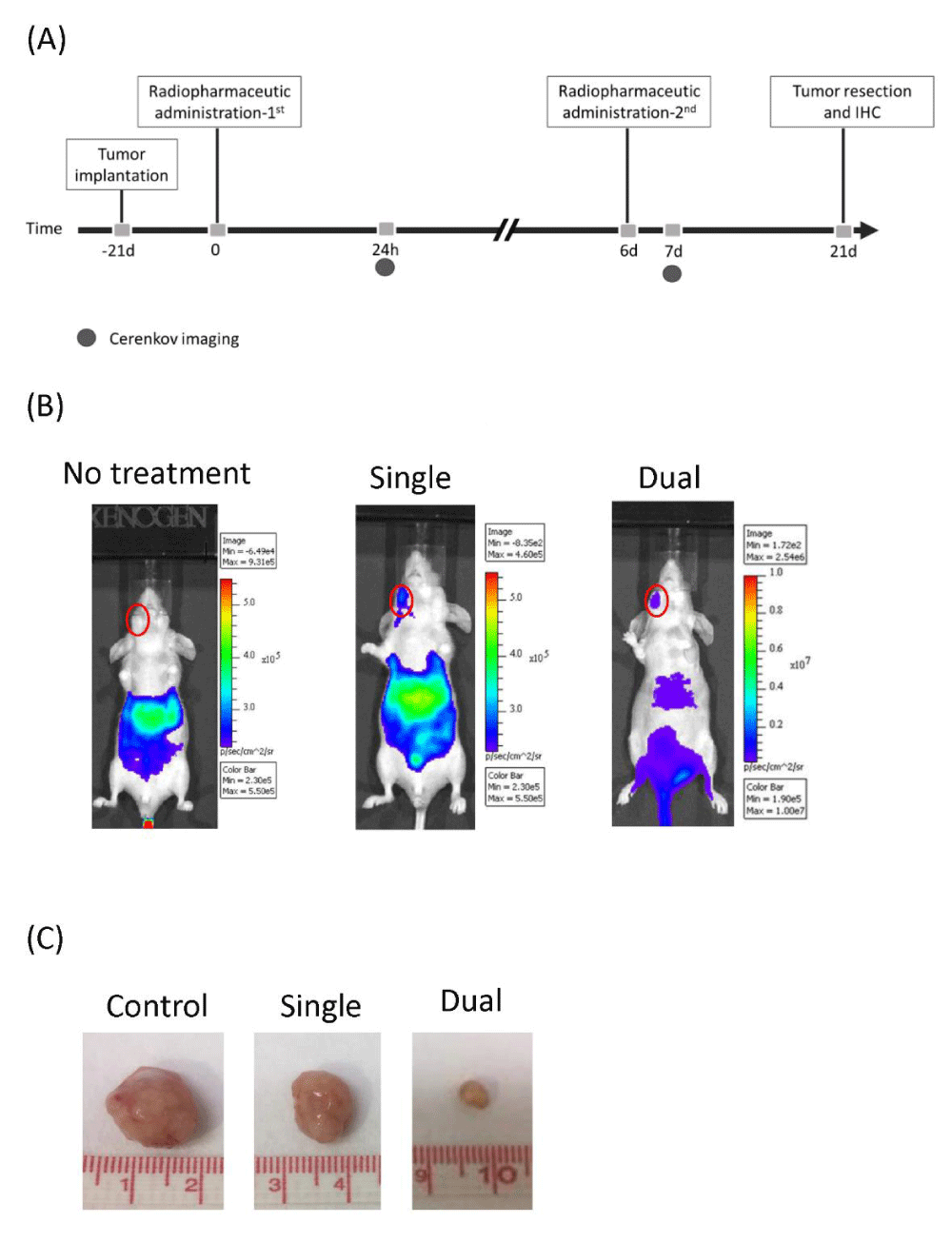
The regimen of 188Re-liposome treatment and the timeline of imaging evaluation and tumor resection on HNSCC tumor-bearing mice were schemed (Figure 1A). Because 188Re-liposome emits high energy β-particles, the drug accumulation at tumor site can be detected by the Cerenkov luminescent imaging (CLI) in vivo. Compared to the untreated control, both strategies of 188Re-liposome treatment showed CLI signals in tumor-bearing mice (Figure 1B). Although dual doses did not exhibit increased accumulation of 188Re-liposome at tumor lesion, the tumor size was apparently smaller than that treated with a single dose of 188Re-liposome (Figure 1C). These results implied that dual doses of 188Re-liposome would be more effective than a single dose of 188Re-liposome on suppression of tumor growth in vivo.
Figure 1: Comparison of dual doses and a single dose of 188Re-liposome on suppression of orthotopic HNSCC tumor model. (A) The experimental scheme of 188Re-liposome treatment and tumor evaluation. (B) Cerenkov luminescent imaging (CLI) of orthotopic tumor treated with 188Re-liposome. (C) Comparison of sizes of the resected tumors from tumor-bearing mice with or without 188Re-liposome treatment.
Effects of single dose and dual doses of 188Re-liposome on the expression of Ki67 biomarker
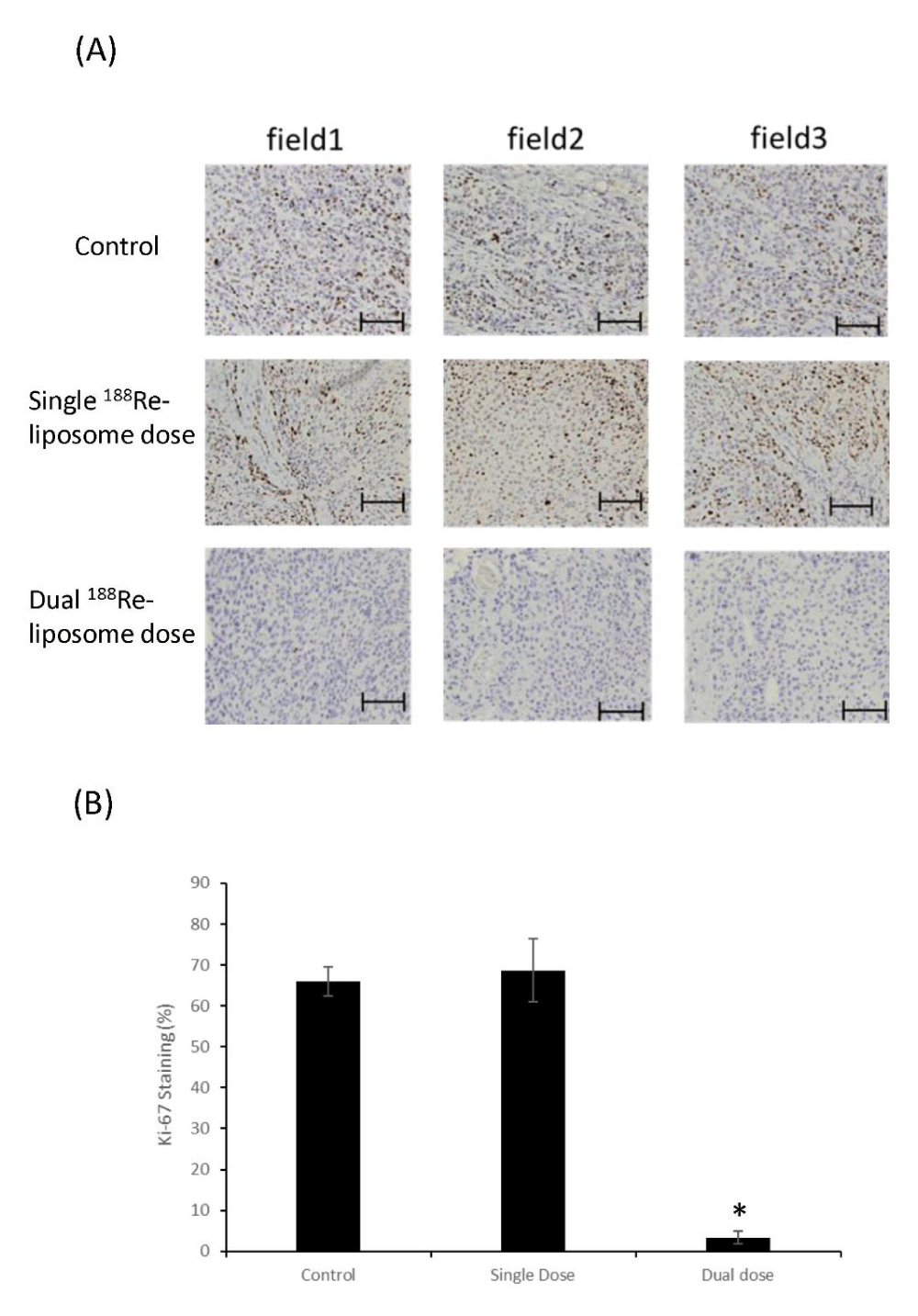
To determine if different effects of tumor suppression by single dose and dual doses of 188Re-liposome was associated with tumor proliferation, the Ki-67 proliferative marker was examined in sections of resected tumors. Compared to the untreated control and the single dose, tumors treated with dual doses of 188Re-liposome expressed a very low level of Ki-67 using IHC staining (Figure 2A). The result was also quantified by IHC scoring (Figure 2B). Therefore, dual doses of 188Re-liposome could better suppress the proliferation of tumor cells in vivo.
Figure 2: Comparison of Ki-67 proliferative marks in HNSCC tumor sections. (A) IHC staining of Ki-67 in tumor sections with or without the treatment of 188Re-liposome. Three random fields were selected for imaging acquisition and quantification. Scale bar: 100 m. (B) Quantification of IHC staining of Ki-67 markers in tumor treated with different regimes of 188Re-liposome. *: p < 0.05.
Effects of single dose and dual doses of 188Re-liposome on the expression of EMT related biomarkers
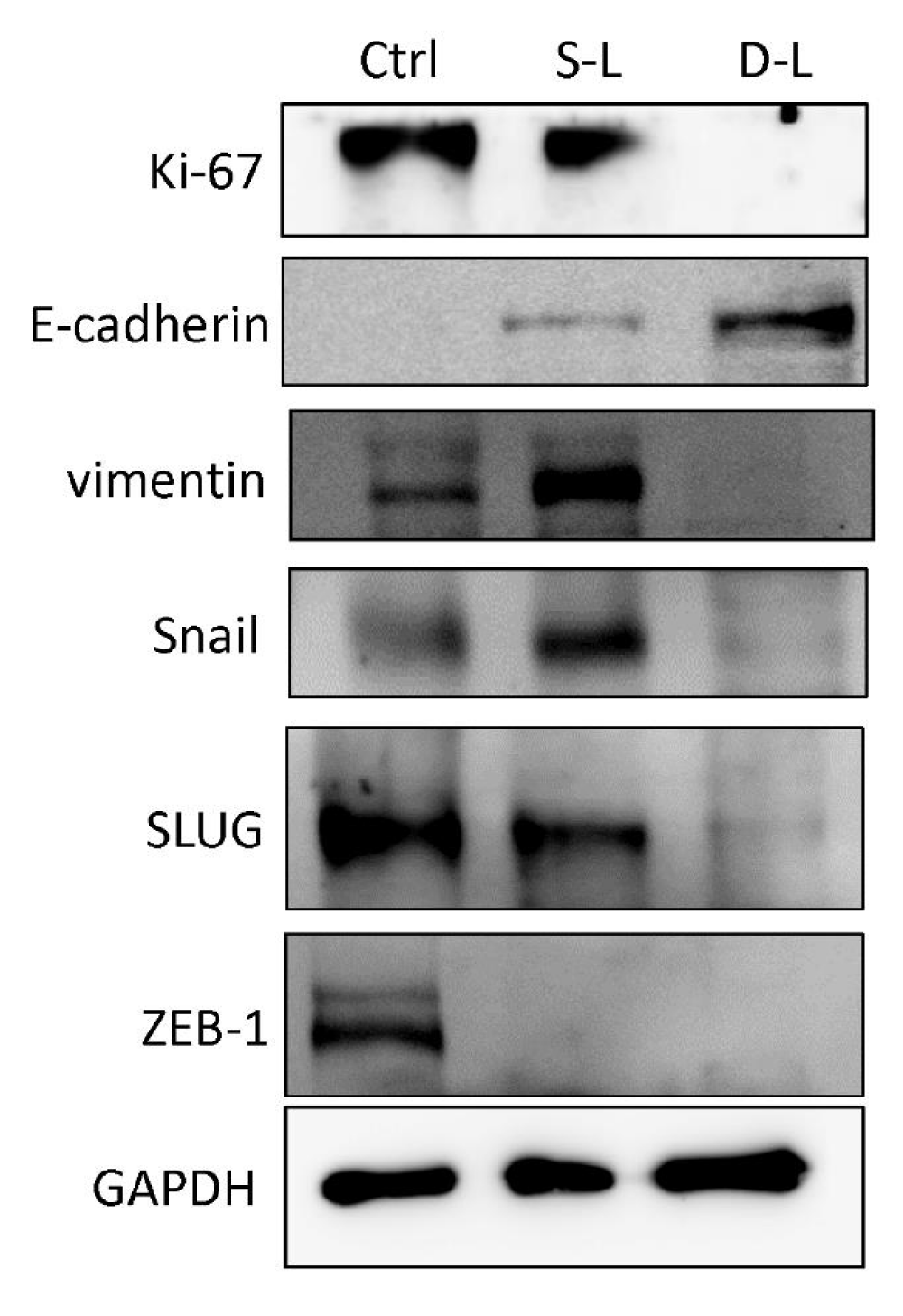
In addition to the Ki-67 proliferative marker, we also compared the expression of biomarkers associated with EMT mechanism in HNSCC tumors treated with a single dose and dual doses of 188Re-liposome. E-cadherin, vimentin, Snail, Slug, and ZEB-1 were examined, and the results showed that dual doses of 188Re-liposome exhibited stronger effects on suppression of EMT by inducing E-cadherin, and inhibiting vimentin, Snail and Slug, respectively (Figure 3). ZEB-1 was the only EMT promoting molecule suppressed equally by both single dose and dual doses of 188Re-liposome (Figure 3). According to the expression of these molecules, it suggests that dual doses of 188Re-liposome enhanced the therapeutic efficacy by suppressing tumor proliferation and metastasis concomitantly.
Figure 3: Effects of 188Re-liposome on the expression of EMT related biomarkers. Western blot analysis was used to detect the protein levels of these markers expressed in HNSCC tumors treated with dual doses or a single dose of 188Re-liposome compared to untreated controls.
Comparison of microRNA expressive profiles in HNSCC tumors treated with a single dose and dual doses of 188Re-liposome
We have previously found that 188Re-liposome can induce let-7 microRNA as elucidated from the microarray analysis [12]. Here we further investigated the expressive profiles of microRNA in HNSCC tumors treated with 188Re-liposome using the Taqman® Openarray for human microRNA. The heatmaps of microRNA open arrays overviewed a total of 758 microRNA in HNSCC tumors treated with a single dose of 188Re-liposome, dual doses of 188Re-liposome, and untreated control (Supplementary data 1). We shortlisted a group of the most up-regulated and down-regulated miRNAs who have opposite expression profiles between dual doses and a single dose of 188Re-liposome treatment. Cut-off expressive ratios (in log2) between dual doses and a single dose treatment (D/S ratio) were set to be 2 and -1 for up- and down-regulation groups, respectively. Twenty-two microRNAs (miRBase ID: miR-181c-5p, miR-338-5p, miR-1285-3p, miR-146a-5p, miR-25-5p, miR-1225-3p, miR-147b, miR-28-3p, miR-10b-3p, miR-99b-3p, miR-577, miR-361-3p, miR-1260a, miR-150-5p, miR-148b-3p, miR-186-5p, miR-543, miR-592, miR-501-3p, miR-23b-3p, miR-766-3p, and miR-342-3p) were up-regulated by dual doses of 188Re-liposome but concomitantly down-regulated by a single dose of 188Re-liposome (Figure 4A). On the other hand, nineteen microRNAs (miR-872, miR-200a-5p, miR-1267, miR-296-5p, miR-584-5p, miR-29a-5p, miR-200c-5p, miR-1233-5p, miR-let-7i-3p, miR-21-3p, miR-522-3p, miR-629-5p, miR-18a-3p, miR-520e, miR-224-5p, miR-200b-5p, miR-208b-3p, miR-744-5p, and miR-551b-5p) were down-regulated by dual doses of 188Re-liposome accompanied by up-regulation with a single dose of 188Re-liposome (Figure 4B). These two microRNA groups were separately subjected to the KEGG pathway analysis. Most of the genes affected by microRNAs that were up-regulated or down-regulated by dual doses of 188Re-liposome were associated with pathways in cancer as well as other cancer-associated pathways (Table 1 and Table 2). These results suggest that 188Re-liposome would regulate the expression of microRNA in HNSCC and ablate tumor progression.
Supplementary data 1: The overall heatmaps of Taqman Openarrays microRNA panel revealed the expressive profiles of 758 microRNA in HNSCC Tumors treated with a single dose of 188Re-liposome, dual dose of 188Re-liposome, and untreated control.
Figure 4: Comparison of microRNAs inversely regulated by different treatment regimes of 188Re-liposome on the orthotopic HNSCC tumor model. (A) The microRNAs up-regulated by dual doses and concomitantly down-regulated by a single dose of 188Re-liposome. (B) The microRNAs down-regulated by dual doses and concomitantly up-regulated by a single dose of 188Re-liposome. The differences between dual doses and single dose of 188Re-liposome regulated microRNA expression (log2) were represented by D/S ratio of microRNA./p>
| Table 1: Top 10 pathways associated with miRNAs down-regulated by dual doses but up-regulated by single dose of 188Re-liposome | |||
| KEGG pathway | #genes | #miRNAs | p - value |
| Pathways in cancer | 107 | 14 | 0.03296646 |
| MAPK signaling pathway | 81 | 15 | 0.00192409 |
| Proteoglycans in cancer | 67 | 14 | 6.89E-08 |
| Ras signaling pathway | 63 | 15 | 0.01942799 |
| Rap1 signaling pathway | 60 | 12 | 0.01508721 |
| Transcriptional misregulation in cancer | 55 | 14 | 0.02290762 |
| Hippo signaling pathway | 50 | 11 | 6.89E-08 |
| Protein processing in endoplasmic reticulum | 47 | 12 | 0.0434422 |
| Signaling pathways regulating pluripotency of stem cells | 46 | 15 | 0.00030753 |
| Dopaminergic synapse | 45 | 12 | 0.00809897 |
| Table 2: Top 10 pathways associated with miRNAs up-regulated by dual doses but down-regulated by single dose of 188Re-liposome. | |||
| KEGG pathway | #genes | #miRNAs | p - value |
| Pathways in cancer | 175 | 21 | 0.025728198 |
| PI3K-Akt signaling pathway | 153 | 21 | 0.002721846 |
| MAPK signaling pathway | 120 | 19 | 0.002721846 |
| Ras signaling pathway | 104 | 19 | 0.002721846 |
| Regulation of actin cytoskeleton | 102 | 19 | 0.010643911 |
| Rap1 signaling pathway | 101 | 21 | 0.001949161 |
| Proteoglycans in cancer | 100 | 19 | 0.00015307 |
| Focal adhesion | 97 | 19 | 0.009082868 |
| Endocytosis | 94 | 18 | 0.018303813 |
| cAMP signaling pathway | 90 | 20 | 0.029635054 |
| cGMP-PKG signaling pathway | 82 | 20 | 0.002742686 |
Demonstration of potent microRNA regulated by single dose and dual doses of 188Re-liposome
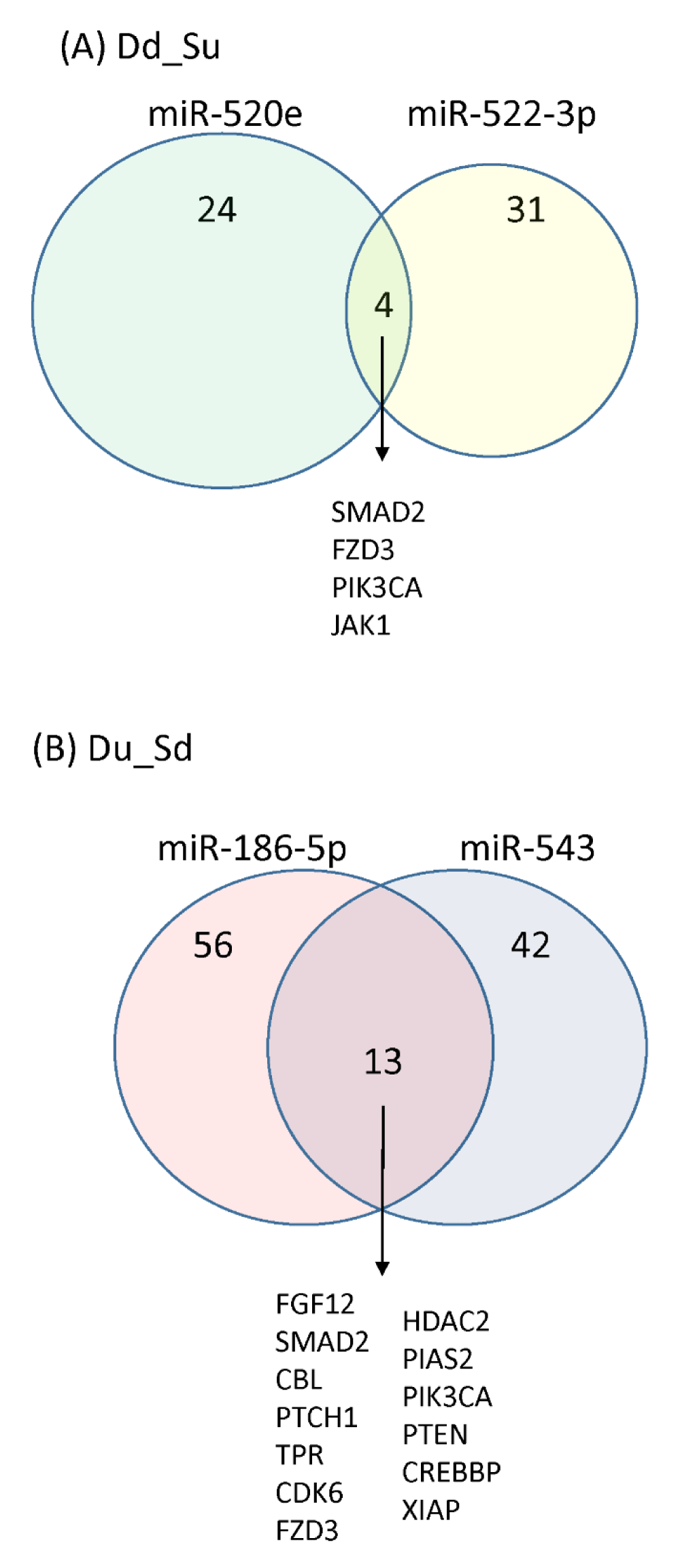
According to the KEGG pathway analysis by the microRNA openarray dataset, the intracellular signaling pathways of HNSCC tumors influenced by dual doses of 188Re-liposome were ranked by p - value. Thirty-nine and 62 pathways were significantly affected by the microRNAs down-regulated and up-regulated by dual doses of 188Re-liposome, respectively (p < 0.05). According to the rank of p - value, the top 10 signaling pathways affected by miRNAs that were down-regulated or up-regulated by dual doses of 188Re-liposome were different and were shown in table 3 and table 4. Compared to other microRNAs down-regulated by dual doses of 188Re-liposome, miR-520e and miR-522-3p affected most of the genes involved in that top 10 pathways (Table 3). On the other hand, miR-186-5p and miR-543 were involved in another 10 signaling pathways mostly affected by dual doses of 188Re-liposome up-regulated miRNAs (Table 4). We next compared the downstream genes regulated by miR-520e and miR-522-3p and found that SMAD2, FZD3, PIK3CA, and JAK1 genes were involved in these two microRNAs (Figure 5A). For miR-186-5p and miR-543, 13 genes including FGF12, SMAD2, CBL, PTCH1, TPR, CDK6, FZD3, HDAC2. PIAS2, PIK3CA, PTEN, CREBBP, and XIAP were co-regulated (Figure 5B). Surprisingly, SMAD2, FZD3, and PIK3CA genes were commonly regulated by these four miRNAs, even though miR520e and miR-522-3p were down-regulated, but miR-186-5p and miR-543 were up-regulated by dual doses of 188Re-liposome. The full list of downstream genes regulated by these four microRNAs was provided (Supplementary data 2).
Figure 5: The Venn diagram of different microRNAs regulated downstream genes regulated by different regimes of 188Re-liposome. (A) A collection of downstream genes from miR-520e and miR-522-3p down-regulated by dual doses of 188Re-liposome. (B) A collection of downstream genes from miR-186-5p and miR-543 down-regulated by dual doses of 188Re-liposome.
Supplementary data 2: The downstream genes regulated by 188Re-liposome regulated microRNAsa.
| Table 3: Dual doses of 188Re-liposome down-regulated miRNAs that influence most genes and their associated KEGG pathways. | ||||||
| KEGG pathway | hsa-miR-520e (774 genes in db) | hsa-miR-522-3p (1069 genes in db) | ||||
| genes involved | % Involvement | % target genes in db | genes involved | % Involvement | % target genes in db | |
| Hippo signaling pathway | 9 | 18% | 1.163% | 23 | 46% | 2.152% |
| Proteoglycans in cancer | 18 | 27% | 2.326% | 18 | 27% | 1.684% |
| Lysine degradation | 6 | 33% | 0.775% | 4 | 22% | 0.374% |
| TGF-beta signaling pathway | 11 | 41% | 1.421% | 10 | 37% | 0.935% |
| Signaling pathways regulating pluripotency of stem cells | 13 | 28% | 1.680% | 17 | 37% | 1.590% |
| Mucin type O-Glycan biosynthesis | 2 | 20% | 0.258% | 4 | 40% | 0.374% |
| N-Glycan biosynthesis | 2 | 15% | 0.258% | 2 | 15% | 0.187% |
| Morphine addiction | 3 | 11% | 0.388% | 8 | 30% | 0.748% |
| Glioma | 6 | 25% | 0.775% | 6 | 25% | 0.561% |
| MAPK signaling pathway | 23 | 28% | 2.972% | 23 | 28% | 2.152% |
| Table 4: Dual doses of 188Re-liposome up-regulated miRNAs that influence most genes and their associated KEGG pathways. | ||||||
| KEGG pathway | hsa-miR-186-5p (1649 genes in db) | hsa-miR-543 (1556 genes in db) | ||||
| genes involved | % Involvement | % target genes in db | genes involved | % Involvement | % target genes in db | |
| Prion diseases | 6 | 38% | 0.364% | 3 | 19% | 0.193% |
| ECM-receptor interaction | 7 | 18% | 0.424% | 5 | 13% | 0.321% |
| Adrenergic signaling in cardiomyocytes | 29 | 36% | 1.759% | 18 | 23% | 1.157% |
| Hippo signaling pathway | 26 | 37% | 1.577% | 22 | 31% | 1.414% |
| Sphingolipid signaling pathway | 19 | 29% | 1.152% | 19 | 29% | 1.221% |
| Adherens junction | 12 | 27% | 0.728% | 15 | 33% | 0.964% |
| Thyroid hormone signaling pathway | 20 | 32% | 1.213% | 19 | 30% | 1.221% |
| Estrogen signaling pathway | 18 | 38% | 1.092% | 13 | 28% | 0.835% |
| Glioma | 17 | 47% | 1.031% | 12 | 33% | 0.771% |
| Amphetamine addiction | 21 | 54% | 1.273% | 16 | 41% | 1.028% |
Association of miRNAs affected by 188Re-liposome and patients survival
We next analyzed the association of miR-520e, miR-522-3p, miR-186-5p, and miR-543 with patients survival using the public on-line Kaplan-Meier Plotter tool (see Materials and Methods). High expression of miR-520e and miR-522-3p was associated with a reduced survival rate, while that of miR-186-5p was marginally associated with an increased survival rate of HNSCC cancer patients (Figure 6). On the other hand, higher expression of miR-543 was also associated with lower survival rate (data not shown). As mentioned above, miR-520e and miR-522-3p were suppressed by dual doses of 188Re-liposome treatment, but miR-186-5p was up-regulated accordingly. Hence, it may suggest that these microRNAs could be used as prognostic factors for HNSCC patients treated with 188Re-liposome.
Figure 6: The survival analysis of 188Re-liposome regulated microRNAs using HNSCC patient database. Comparison of high and low expression of miR-520e, miR-522, and miR-186 on overall survivals of on HNSCC patient using the Kaplan-Meier statistics with a log-rank test. p < 0.05 represented a significant difference.
188Re-liposome has been demonstrated to be effective on suppression a variety of human cancers. Most of the studies focused on the biodistribution, tumor targeting, pharmaceutic kinetics and dosimetry previously [9,10,17,22-24]. A phase 0 clinical trial has also shown that 188Re-liposome is a potent theranostic agent for cancer treatment [25]. In this study, we first demonstrated that dual doses of 188Re-liposome exhibited stronger effects than a single dose of 188Re-liposome on suppression of orthotopic HNSCC tumor growth and EMT phenomenon. We have recently showed that repeated treatment of 188Re-liposome on tumor-bearing mice do not cause acute toxicity but reduce blood cell counts [15]. The time interval between the two administrations of 188Re-liposome was over 10 half-lives of this isotope, yet the responses of HNSCC tumors were still enhanced. It is speculated that tumor insulted by first dose of 188Re-liposome has not been fully repaired before the second doses of 188Re-liposome. Notably, the dose of dual treatments were equal and both were at 80% MTD. Thus a sublethal damage repair should not be raised to compromise the efficacy of 188Re-liposome at second treatment.
Most of microRNAs down-regulated by dual doses of 188Re-liposome but up-regulated by a single dose of 188Re-liposome, or vice versa, were associated with genes involved in pathways in cancer according to the KEGG pathway analysis. Although the p - value of this category was not smallest in affected pathways (Table 1 and 2), it is convinced that dual doses of 188Re-liposome would vigorously influence microRNA related genes in tumor ablation. Interestingly, the pathway of proteoglycan in cancer accounted for the smallest p - value in both conditions of microRNA regulated by dual doses of 188Re-liposome. The signaling of hyaluronan, heparan sulfate proteoglycans, chondroitin sulfate/dermatan sulfate proteoglycan, and keratan sulfate proteoglycan are involved in proteoglycan in cancer pathway, and they are essential for cell adhesion, migration and angiogenesis in KEGG pathway. Indeed, the role of proteoglycan in tumor microenvironment and angiogenesis has been reported [26-28]. Thus, this bioinformatics analysis for microRNA regulation was consistent with the tumor suppressive effects caused by dual doses of 188Re-liposome. The p - value of Hippo signaling pathway was the same with proteoglycan in cancer pathway only in microRNAs down-regulated by dual doses of 188Re-liposome. This pathway was even not ranked as primary pathway affected by 188Re-liposome up-regulated microRNAs (Table 2). As Hippo signaling is a critical tumor suppressor pathway, it would be interesting to further investigate how 188Re-liposome regulate this signaling pathway.
According to the data analysis of Taqman® Openarray Human MicroRNA Panel, miR-520e and miR-522-3p were down-regulated by dual doses of 188Re-liposome but they were also induced by a single dose of 188Re-liposome. On the contrary, miR-186-5p and miR-543 were regulated oppositely by the same regime. Although these microRNAs did not rank as top change by dual doses over a single dose of 188Re-liposome, they influenced most genes in top 10 pathways affected by dual doses of 188Re-liposome. The Venn diagram showed that miR-520e and miR-522-3p co-regulated 4 downstream genes, and miR-186-5p and miR-543 co-regulated 13 downstream genes. Surprisingly, these two groups of microRNAs responded oppositely to dual doses of 188Re-liposome influenced 3 common genes, that is, SMAD2, FZD3, and PIK3CA. SMAD2 mediates transforming growth factor-β (TGF-β) relayed signaling pathway, and it works differentially with other SMAD isoform [29]. TGF-β signaling pathway contains both tumor suppressor and oncogene actions mediated by SMAD3 [30]. However, several lines of evidence showed that SMAD2 belongs to tumor suppressor gene of TGF-β signaling pathway [31,32]. Little is known about the function of FZD3 (Frizzled 3 receptor) gene, although a recent report demonstrated that down-regulation of FZD3 gene suppresses human melanoma tumorigenesis independent of WNT signaling [33]. PIK3CA (Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha) gene is also regarded an oncogene, and is usually mutated in cancer cells [34]. It is believed that other genes independently regulated by these two groups of microRNAs are also associated with tumorigenesis. Although we focused on these four microRNAs up-regulated or down-regulated by dual doses of 188Re-liposome, other microRNAs are not excluded and further investigation should be required.
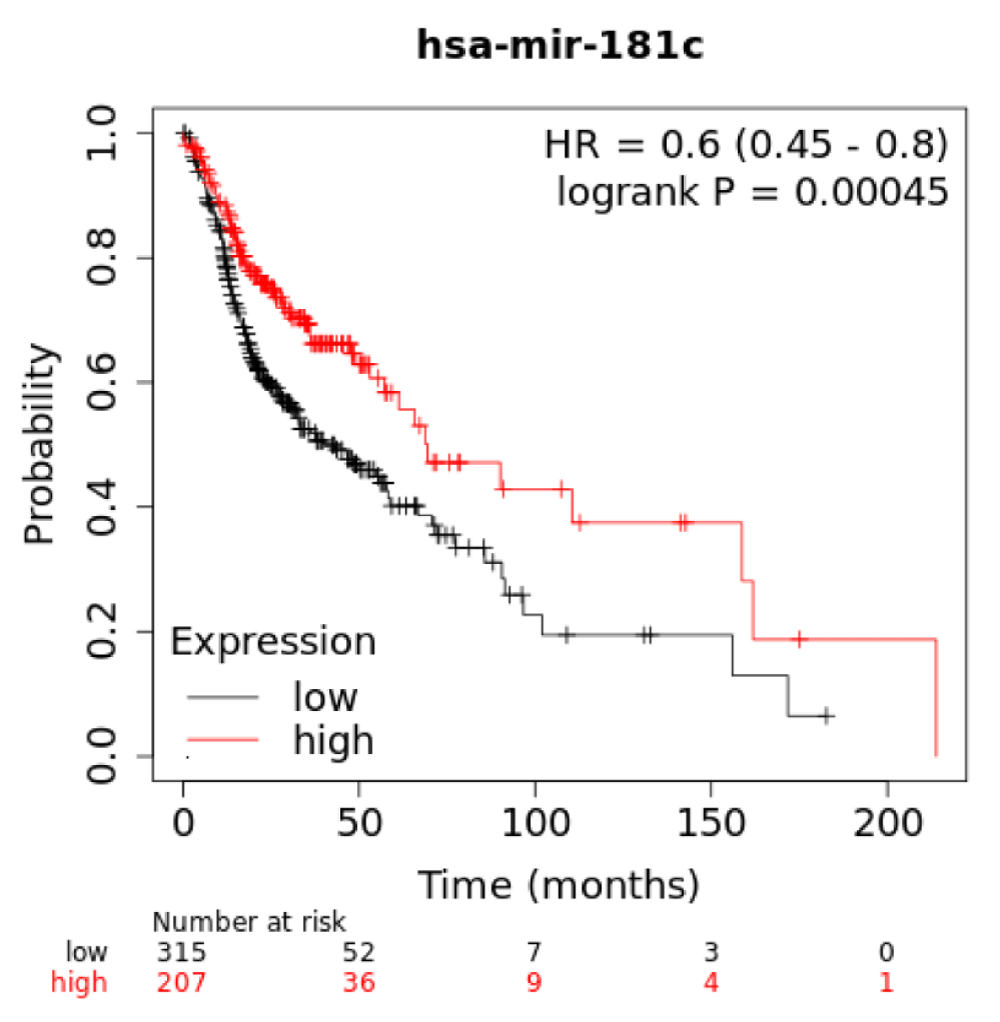
For clinical relevant, we applied the Kaplan-Meier survival analysis to examine if the 188Re-liposome regulated microRNA would be associated with the survival rate of HNSCC patients. For dual doses of 188Re-liposome down-regulated miR-520e and miR-522-3p, both of them exhibit reduced survival rates at high expression. Notably, these two microRNAs were up-regulated by a single dose of 188Re-liposome. Dual doses of 188Re-liposome up-regulated miR-186-5p was associated with enhanced survival rate at high expression, but the significance is margin. MiR-181c-5p was ranked as highest D/S ratio, and this microRNA exhibited significant increase of survival rate at high expression (Supplementary data 3). However, miR-872 had the lowest D/S ratio but the association of this microRNA with survival rate was unavailable using the online KM plot tool. Although current study could not determine the biological significance of these microRNA in modulating the therapeutic efficacy of 188Re-liposome, they might be considered as prognostic factors for different regime of 188Re-liposome treatment.
Supplementary data 3: Association of miR-181c and HNSCC patient survival.
Current data demonstrated that dual doses of 188Re-liposome exhibited better tumor ablation than a single dose of 188Re-liposome on the orthotopic HNSCC tumor model. We further found that several microRNAs were inversely regulated by these two regimes of 188Re-liposome treatment. The bioinformatics analysis showed that these microRNAs (41 in total) were mainly involved in cancer related pathways. The specific microRNAs found to be involved in regulation of most of downstream genes in dual doses of 188Re-liposome influenced signaling pathways were associated with survival rates of HNSCC patients based on the public microRNA database. For instance, miR-520e and miR-522-3p down-regulated by dual doses but up-regulated by a single dose of 188Re-liposome were associated with worse survival rate when both of them highly expressed in HNSCC patients. Although the role of these microRNAs on mediating the efficacy of 188Re-liposome on HNSCC tumor remains obscure, they might be used as prognostic factors for evaluating different regimes of 188Re-liposome treatment, at least in part.
This work was supported by National Yang-Ming University-Far Eastern Memorial Hospital Joint Research Program (107DN03 and 110DN07), and a grant from Ministry of Science and Technology (MOST 109-2314-B-010-021-MY3). We thank Institute of Nuclear and Energy Research of Taoyuan, Taiwan produced 188Re for preparation of 188Re-liposome. We thank the Taiwan Animal Consortium (MOST 108-2319-B-001-003)--Taiwan Mouse Clinic which is funded by the Ministry of Science and Technology (MOST) of Taiwan for technical support in in vivo imaging experiments. We also thank the support from the Cancer Progression Research Center, National Yang-Ming University, from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
- Bodalal Z, Trebeschi S, Nguyen-Kim TDL, Schats W, Beets-Tan R. Radiogenomics: bridging imaging and genomics. Abdom Radiol (NY). 2019; 44: 1960-1984. PubMed: https://pubmed.ncbi.nlm.nih.gov/31049614/
- Snyder AR, Morgan WF. Gene expression profiling after irradiation: clues to understanding acute and persistent responses? Cancer Metastasis Rev. 2004; 23: 259-268. PubMed: https://pubmed.ncbi.nlm.nih.gov/15197327/
- Yordanova A, Eppard E, Kurpig S, Bundschuh RA, Schonberger S, et al. Theranostics in nuclear medicine practice. Onco Targets Ther. 2017; 10: 4821-4828. PubMed: https://pubmed.ncbi.nlm.nih.gov/29042793/
- Lepareur N, Lacoeuille F, Bouvry C, Hindre F, Garcion E, et al. Rhenium-188 Labeled Radiopharmaceuticals: Current Clinical Applications in Oncology and Promising Perspectives. Front Med (Lausanne). 2019; 6: 132. PubMed: https://pubmed.ncbi.nlm.nih.gov/31259173/
- Kennedy CM, Pinkerton TC. Technetium carboxylate complexes--I. A review of Tc, Re and Mo carboxylate chemistry. International journal of radiation applications and instrumentation Part A, Applied radiation and isotopes. 1988; 39: 1159-1165.
- Deutsch E, Libson K, Vanderheyden JL, Ketring AR, Maxon HR. The chemistry of rhenium and technetium as related to the use of isotopes of these elements in therapeutic and diagnostic nuclear medicine. Int J Rad Appl Instrum B. 1986; 13: 465-477. PubMed: https://pubmed.ncbi.nlm.nih.gov/3793504/
- Wang HY, Lin WY, Chen MC, Lin T, Chao CH, et al. Inhibitory effects of Rhenium-188-labeled Herceptin on prostate cancer cell growth: a possible radioimmunotherapy to prostate carcinoma. Int J Radiation Biol. 2013; 89: 346-355.
- Chang CH, Liu SY, Chi CW, Yu HL, Chang TJ, et al. External beam radiotherapy synergizes (1)(8)(8)Re-liposome against human esophageal cancer xenograft and modulates (1)(8)(8)Re-liposome pharmacokinetics. Int J Nanomedicine. 2015; 10: 3641-3649. PubMed: https://pubmed.ncbi.nlm.nih.gov/26056445/
- Lin LT, Chang CH, Yu HL, Liu RS, Wang HE, et al. Evaluation of the therapeutic and diagnostic effects of PEGylated liposome-embedded 188Re on human non-small cell lung cancer using an orthotopic small-animal model. J Nucl Med. 2014; 55: 1864-1870. PubMed: https://pubmed.ncbi.nlm.nih.gov/25349220/
- Huang FY, Lee TW, Chang CH, Chen LC, Hsu WH, et al. Evaluation of 188Re-labeled PEGylated nanoliposome as a radionuclide therapeutic agent in an orthotopic glioma-bearing rat model. Int J Nanomedicine. 2015; 10: 463-473. PubMed: https://pubmed.ncbi.nlm.nih.gov/25624760/
- Chang YJ, Chang CH, Yu CY, Chang TJ, Chen LC, et al. Therapeutic efficacy and microSPECT/CT imaging of 188Re-DXR-liposome in a C26 murine colon carcinoma solid tumor model. Nucl Med Biol. 2010; 37: 95-104. PubMed: https://pubmed.ncbi.nlm.nih.gov/20122674/
- Lin LT, Chang CY, Chang CH, Wang HE, Chiou SH, et al. Involvement of let-7 microRNA for the therapeutic effects of Rhenium-188-embedded liposomal nanoparticles on orthotopic human head and neck cancer model. Oncotarget. 2016; 7: 65782-6596. PubMed: https://pubmed.ncbi.nlm.nih.gov/27588466/
- Goins B, Bao A, Phillips WT. Techniques for loading technetium-99m and rhenium-186/188 radionuclides into pre-formed liposomes for diagnostic imaging and radionuclide therapy. Methods Mol Biol. 2010; 606: 469-491.
- Maruyama K. Intracellular targeting delivery of liposomal drugs to solid tumors based on EPR effects. Adv Drug Deliv Rev. 2011; 63: 161-169. PubMed: https://pubmed.ncbi.nlm.nih.gov/20869415/
- Chang CY, Chen CC, Lin LT, Chang CH, Chen LC, et al. PEGylated liposome-encapsulated rhenium-188 radiopharmaceutical inhibits proliferation and epithelial-mesenchymal transition of human head and neck cancer cells in vivo with repeated therapy. Cell Death Discov. 2018; 4: 100. PubMed: https://pubmed.ncbi.nlm.nih.gov/30393570/
- Zheng HC. The molecular mechanisms of chemoresistance in cancers. Oncotarget. 2017; 8: 59950-59964. PubMed: https://pubmed.ncbi.nlm.nih.gov/28938696/
- Chang YJ, Chang CH, Chang TJ, Yu CY, Chen LC, et al. Biodistribution, pharmacokinetics and microSPECT/CT imaging of 188Re-bMEDA-liposome in a C26 murine colon carcinoma solid tumor animal model. Anticancer Res. 2007; 27: 2217-2225. PubMed: https://pubmed.ncbi.nlm.nih.gov/17695506/
- Tuominen VJ, Ruotoistenmaki S, Viitanen A, Jumppanen M, Isola J. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010; 12: R56. PubMed: https://pubmed.ncbi.nlm.nih.gov/20663194/
- Tsai CH, Lin LT, Wang CY, Chiu YW, Chou YT, et al. Over-expression of cofilin-1 suppressed growth and invasion of cancer cells is associated with up-regulation of let-7 microRNA. Biochim Biophys Acta. 2015; 1852: 851-861. PubMed: https://pubmed.ncbi.nlm.nih.gov/25597880/
- Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015; 43: W460-466. PubMed: https://pubmed.ncbi.nlm.nih.gov/25977294/
- Nagy A, Lanczky A, Menyhart O, Gyorffy B. Author Correction: Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018; 8: 11515.
- Chen LC, Chang YJ, Chen SJ, Lee WC, Chang CH, et al. Imaging, biodistribution and efficacy evaluation of (188)Re-human serum albumin microspheres via intraarterial route in an orthotopic hepatoma model. Int J Radiat Biol. 2017; 93: 477-486. PubMed: https://pubmed.ncbi.nlm.nih.gov/28045339/
- Chang YJ, Hsu CW, Chang CH, Lan KL, Ting G, et al. Therapeutic efficacy of 188Re-liposome in a C26 murine colon carcinoma solid tumor model. Invest New Drugs. 2013; 31: 801-811. PubMed: https://pubmed.ncbi.nlm.nih.gov/23224353/
- Tsai CC, Chang CH, Chen LC, Chang YJ, Lan KL, et al. Biodistribution and pharmacokinetics of 188Re-liposomes and their comparative therapeutic efficacy with 5-fluorouracil in C26 colonic peritoneal carcinomatosis mice. Int J Nanomedicine. 2011; 6: 2607-2619. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3218575/
- Wang SJ, Huang WS, Chuang CM, Chang CH, Lee TW, et al. A phase 0 study of the pharmacokinetics, biodistribution, and dosimetry of (188)Re-liposome in patients with metastatic tumors. EJNMMI Res. 2019; 9: 46. PubMed: https://pubmed.ncbi.nlm.nih.gov/31119414/
- Hammond E, Khurana A, Shridhar V, Dredge K. The Role of Heparanase and Sulfatases in the Modification of Heparan Sulfate Proteoglycans within the Tumor Microenvironment and Opportunities for Novel Cancer Therapeutics. Front Oncol. 2014; 4: 195. PubMed: https://pubmed.ncbi.nlm.nih.gov/25105093/
- Silver DJ, Siebzehnrubl FA, Schildts MJ, Yachnis AT, Smith GM, et al. Chondroitin sulfate proteoglycans potently inhibit invasion and serve as a central organizer of the brain tumor microenvironment. J Neurosci. 2013; 33: 15603-15617.
- Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011; 15: 1013-1031. PubMed: https://pubmed.ncbi.nlm.nih.gov/21155971/
- Kretschmer A, Moepert K, Dames S, Sternberger M, Kaufmann J, et al. Differential regulation of TGF-beta signaling through SMAD2, Smad3 and Smad4. Oncogene. 2003; 22: 6748-6763. PubMed: https://pubmed.ncbi.nlm.nih.gov/14555988/
- Bae DS, Blazanin N, Licata M, Lee J, Glick AB. Tumor suppressor and oncogene actions of TGFbeta1 occur early in skin carcinogenesis and are mediated by Smad3. Mol Carcinog. 2009; 48: 441-453. PubMed: https://pubmed.ncbi.nlm.nih.gov/18942075/
- Singha PK, Pandeswara S, Geng H, Lan R, Venkatachalam MA, et al. Increased Smad3 and reduced SMAD2 levels mediate the functional switch of TGF-beta from growth suppressor to growth and metastasis promoter through TMEPAI/PMEPA1 in triple negative breast cancer. Genes Cancer. 2019; 10: 134-149. PubMed: https://pubmed.ncbi.nlm.nih.gov/31798766/
- Yang J, Wahdan-Alaswad R, Danielpour D. Critical role of SMAD2 in tumor suppression and transforming growth factor-beta-induced apoptosis of prostate epithelial cells. Cancer Res. 2009; 69: 2185-2190. PubMed: https://pubmed.ncbi.nlm.nih.gov/19276350/
- Li C, Nguyen V, Clark KN, Zahed T, Sharkas S, et al. Down-regulation of FZD3 receptor suppresses growth and metastasis of human melanoma independently of canonical WNT signaling. Proc Natl Acad Sci U S A. 2019; 116: 4548-4557. PubMed: https://pubmed.ncbi.nlm.nih.gov/30792348/
- Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006; 94: 455-459. PubMed: https://pubmed.ncbi.nlm.nih.gov/16449998/